paired end sequencing wikipedia
The inner mate distance. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment.
Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis.

. Library preparation protocols -- In short PE protocols attach an adapter. For subsets of the assemblies we integrated these with additional supporting data to confirm and complement the synteny-based adjacencies. Now lets get started.
Pairs come from the ends of the same DNA strand. Then the paired-end reads for species A can be used to optimise the. Six with physical mapping data that anchor.
The differences between PE and MP reads include. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first generation sequencing. Output per run for single-end.
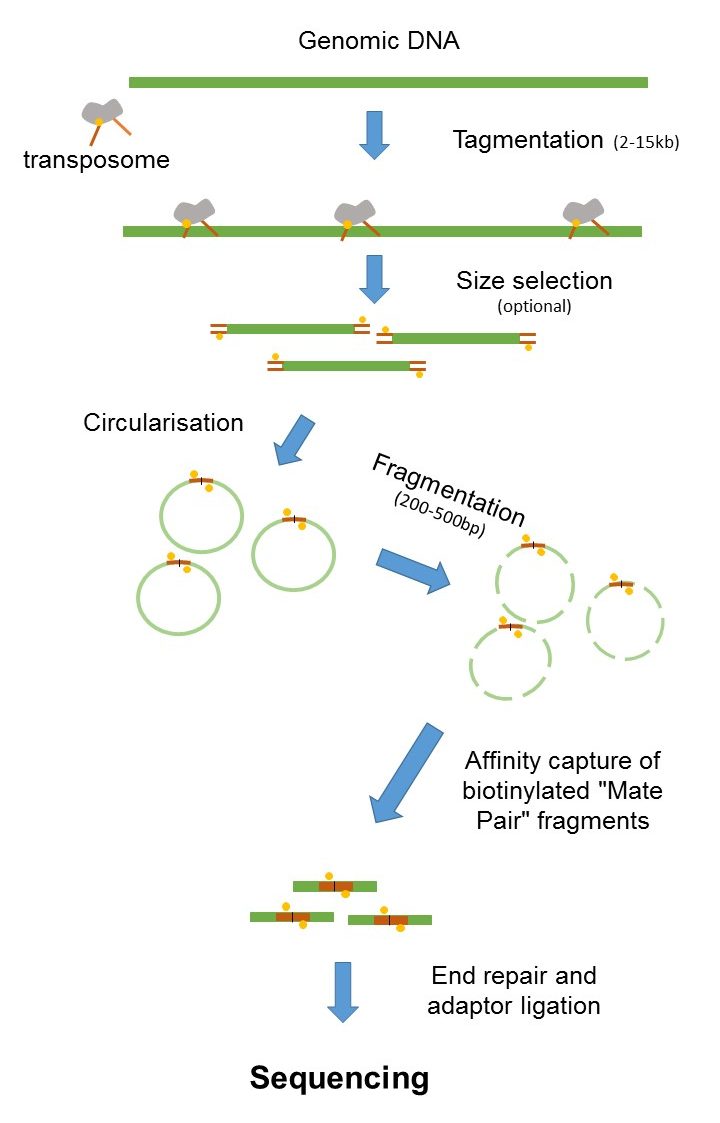
Briefly the target genomic DNA is isolated and partially digested with restrictio. There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.
Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. Any platform that can allow for the ligated fragments to be sequenced across the NheI junction Roche 454 or by paired-end or mate-paired reads Illumina GA and HiSeq platforms would be. A set of paired-end reads must be obtained for species A as part of the genome sequencing project.
Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. This allows the sequencing of both ends of the original template molecule known as paired-end sequencing. End-sequence profiling ESP sometimes Paired-end mapping PEM is a method based on sequence-tagged connectors developed to facilitate de novo genome sequencing to identify high-resolution copy number and structural aberrations such as inversions and translocations.
Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. The present invention provides for a method of preparing a target nucleic acid fragments to produce a smaller nucleic acid which comprises the two ends of the target nucleic acid. Mapping these paired-end reads back to the canonical human genome sequence permits recognition of deletions as pairs of reads that map further apart than expected given.
In conventional paired-end sequencing you simply sequence using the adapter for one end and then once youre done you start over sequencing using the adapter for the other. Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a.
Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. Paired-end sequencing facilitates detection of genomic.
Sequencing information from paired-end reads play an important. Read 1 often called the forward read extends from. Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. The figure shows the. Combining data generated from mate pair.

Rna Seq Basics Applications And Protocol Technology Networks

What Is Mate Pair Sequencing For
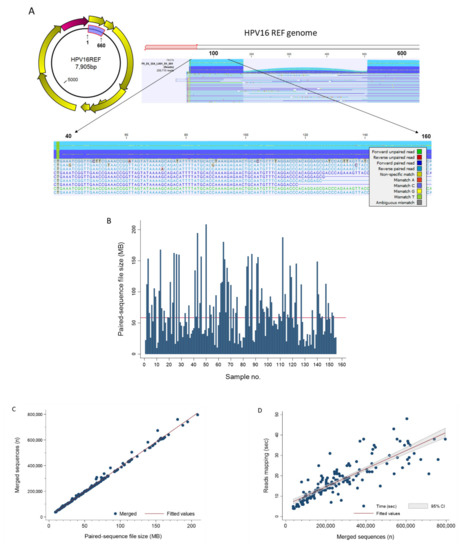
Pathogens Free Full Text Hpv Deepseq An Ultra Fast Method Of Ngs Data Analysis And Visualization Using Automated Workflows And A Customized Papillomavirus Database In Clc Genomics Workbench Html
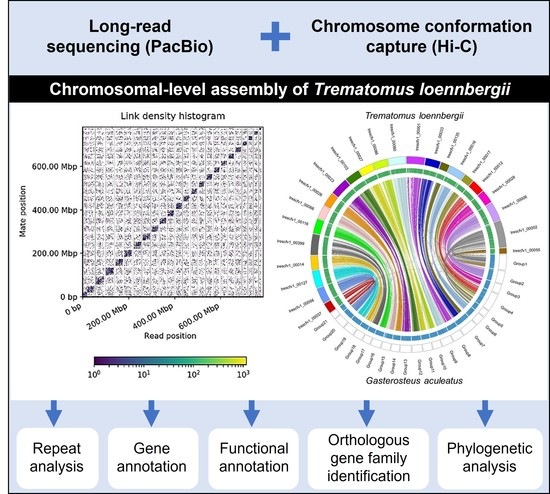
Diversity Free Full Text Chromosomal Level Assembly Of Antarctic Scaly Rockcod Trematomus Loennbergii Genome Using Long Read Sequencing And Chromosome Conformation Capture Hi C Technologies Html
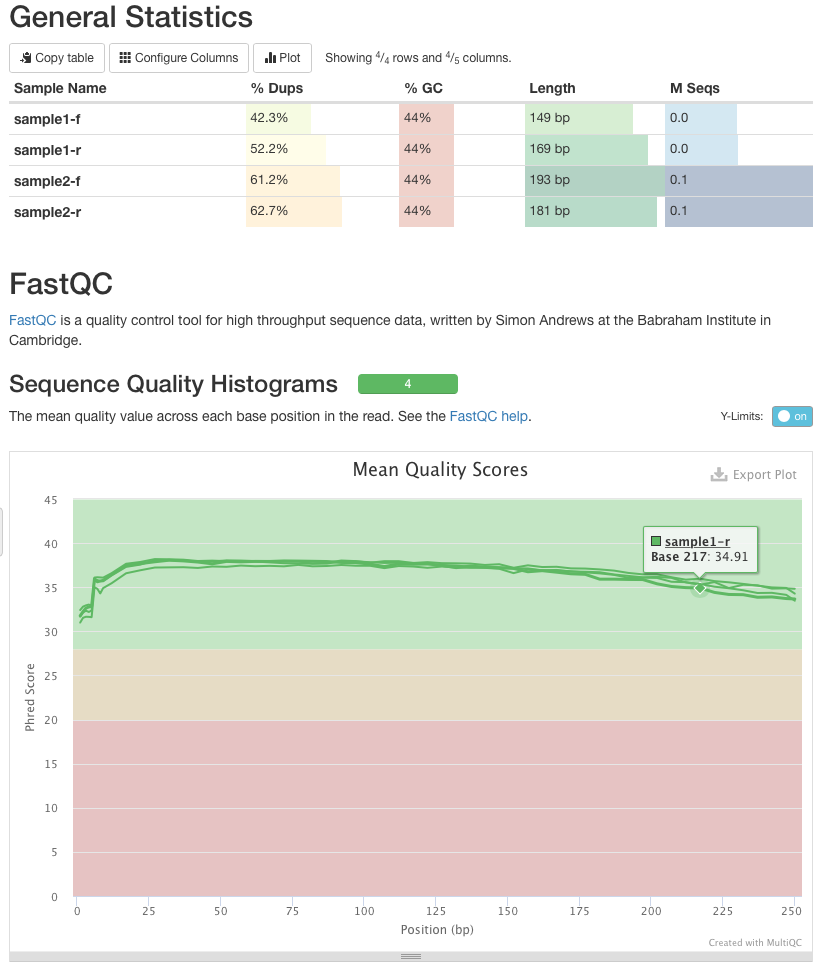
Manipulating Ngs Data With Galaxy Galaxy Community Hub

Bacterial Chromosome Replication Biology Lessons Chromosome Biology
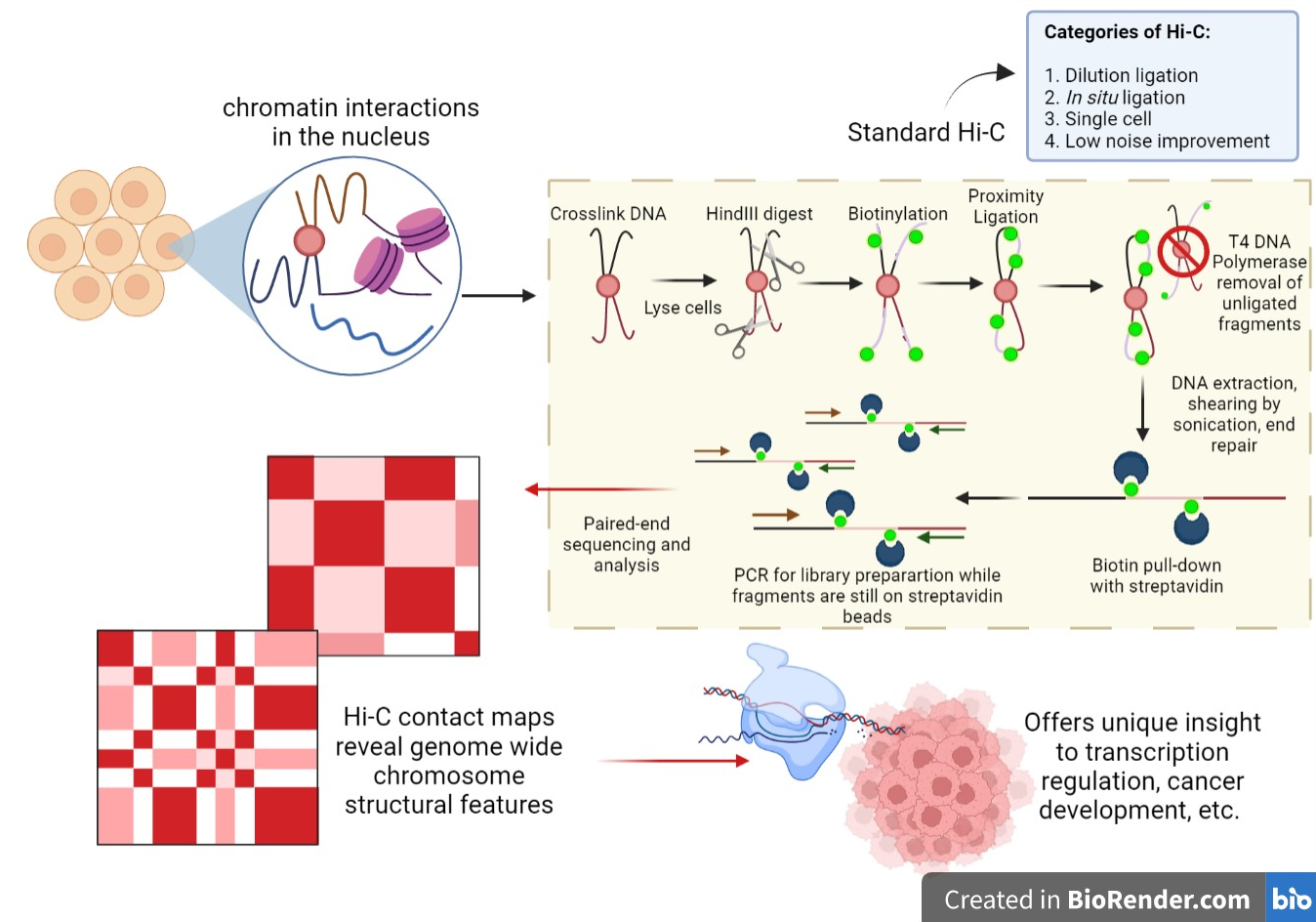
Hi C Genomic Analysis Technique Wikipedia
Informatics For Rna Sequencing A Web Resource For Analysis On The Cloud Plos Computational Biology

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist

Bacterial Chromosome Replication Biology Lessons Chromosome Biology
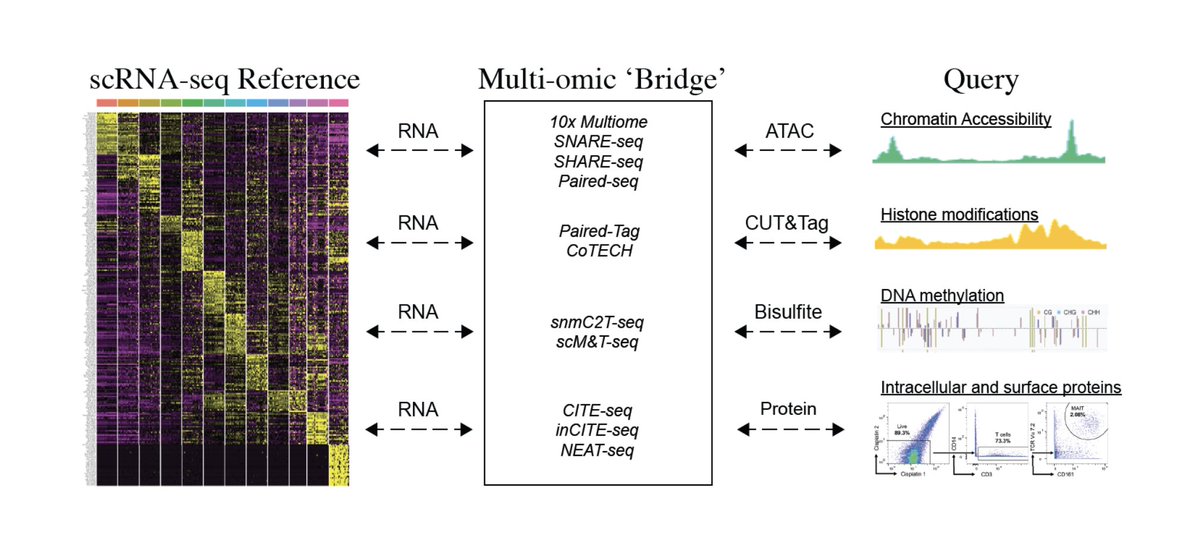
Rahul Satija Satijalab Twitter